Shiny app to create PopArt input files
If you have needed to assemble the input file for PopART this application assists in the creation of the code blocks and the file.
 Available at: Create PopART file
Available at: Create PopART file
about the app
Application is available at the following url Create PopART file
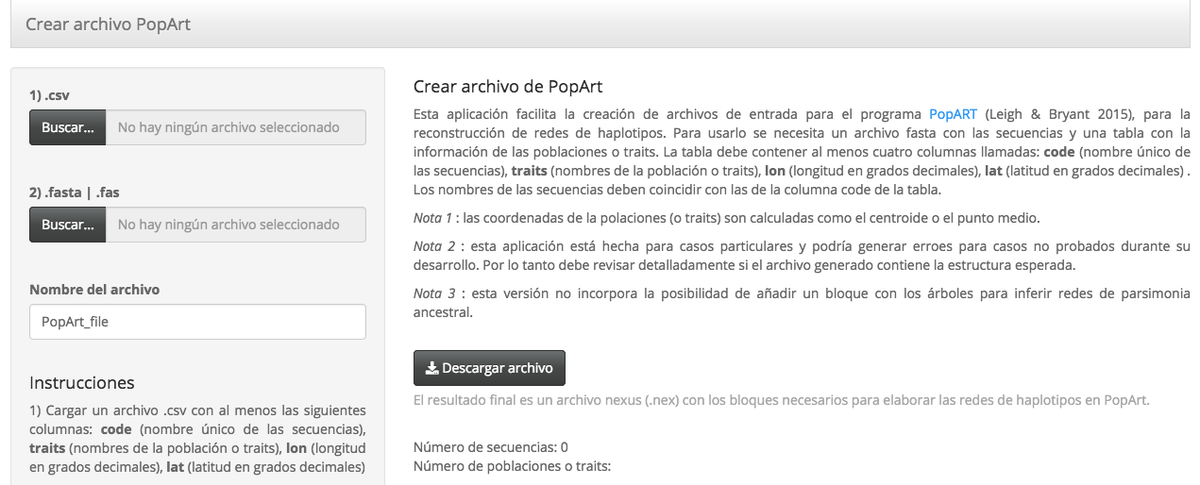
This Shiny application facilitates the creation of PopART input files (Population Analysis with Reticulate Trees; Leigh & Bryant 2015), an haplotype network reconstruction software. To use it, a fasta file with the sequences and a table with the population or trait information are needed. The table must contain at least four columns named: code (unique name of the sequences), traits (population names or traits), lon (longitude in decimal degrees), lat (latitude in decimal degrees). The sequence names must match those in the code column of the table. Please cite the related references.
- Note 1: the coordinates of the poles (or traits) are calculated as the centroid or midpoint.
- Note 2: the application is made for particular cases and could generate errors for cases not tested during its development. Therefore, you should check in detail if the generated file contains the expected structure.
- Note 3: this version does not incorporate the possibility of adding a block with the trees to infer ancestral parsimony networks (to be included).
The final result is a nexus (.nex) file with the necessary blocks to elaborate the haplotype networks in PopArt..
Developed in R with the seqinr, ape, and geosphere packages in Shiny.
This Github repository contains the shiny code and related
Rfunction.
References
Leigh, JW, Bryant D (2015). PopART: Full-feature software for haplotype network construction. Methods Ecol Evol 6(9):1110–1116.